When researchers want to study how COVID makes us sick, or what diseases such as Alzheimer’s do to the body, one approach is to look at what’s happening inside individual cells. Researchers sometimes grow the cells in a 3D scaffold called a “hydrogel.” This network of proteins or molecules mimics the environment the cells would live in inside the body.
New research led by the University of Washington demonstrates a new class of hydrogels that can form not just outside cells, but also inside of them. The team created these hydrogels from protein building blocks designed using a computer to form a specific structure. These hydrogels exhibited similar mechanical properties both inside and outside of cells, providing researchers with a new tool to group proteins together inside of cells.
The team published these results Jan. 30 in the Proceedings of the National Academy of Sciences.
“In the past 10 years, there’s been a shift in the world of cell biology,” said co-senior author Cole DeForest, a UW associate professor of chemical engineering and of bioengineering.
“Classically, folks have attributed much of the cell’s interior organization to membrane-bound organelles, such as mitochondria or the nucleus. But now scientists are realizing that the cell actually has other ways to locally concentrate certain molecules or proteins without using membranes, for example, by protein-protein interactions. This concentrating allows the cell to turn on or off specific functions that can be helpful or ultimately lead to disease.”
DeForest continued, “What I think is pretty exciting here is that we have good mechanical control of our hydrogels—even when they are made inside human cells. This means we can tune them to essentially function as a synthetic version of whatever sequestering phenomenon we want to study, such as how protein aggregation can lead to Alzheimer’s.”
One key element of this research was that the protein building blocks were designed from scratch—they don’t exist anywhere in nature—using computers.
“You can imagine a protein as a string of subunits called amino acids. That string folds up to form a three-dimensional structure. There are 20 different amino acids, and a typical protein is made up of 100 to 200 of them. That makes the system very complex, because how do you know how it’s going to fold?” said co-lead author Rubul Mout, who completed this research as a UW postdoctoral researcher at the Institute for Protein Design and is now a research fellow at Harvard Medical School and Boston Children’s Hospital.
“That’s where the computer comes into play—it does calculations to estimate the most likely three-dimensional shape. And similarly, you can tell it what shape you want and it tells you what sequence you need to build the protein.”
To make a variety of hydrogels with different properties, the team used computational design to control how floppy or rigid the protein building blocks were and how the building blocks organized and connected to create the hydrogel.
The researchers also used two different methods to link the building blocks together: One linked them irreversibly and the other allowed the proteins to disconnect and reconnect.
“Irreversibly crosslinked systems are going to be intrinsically more stable, making them better for long-term cell culture and functional tissue engineering,” said DeForest, who is also a faculty member with the UW Molecular Engineering and Sciences Institute and the UW Institute for Stem Cell and Regenerative Medicine.
“But the reversibly crosslinked systems are more fluid, which may be better for driving specific protein-protein interactions within living cells.”
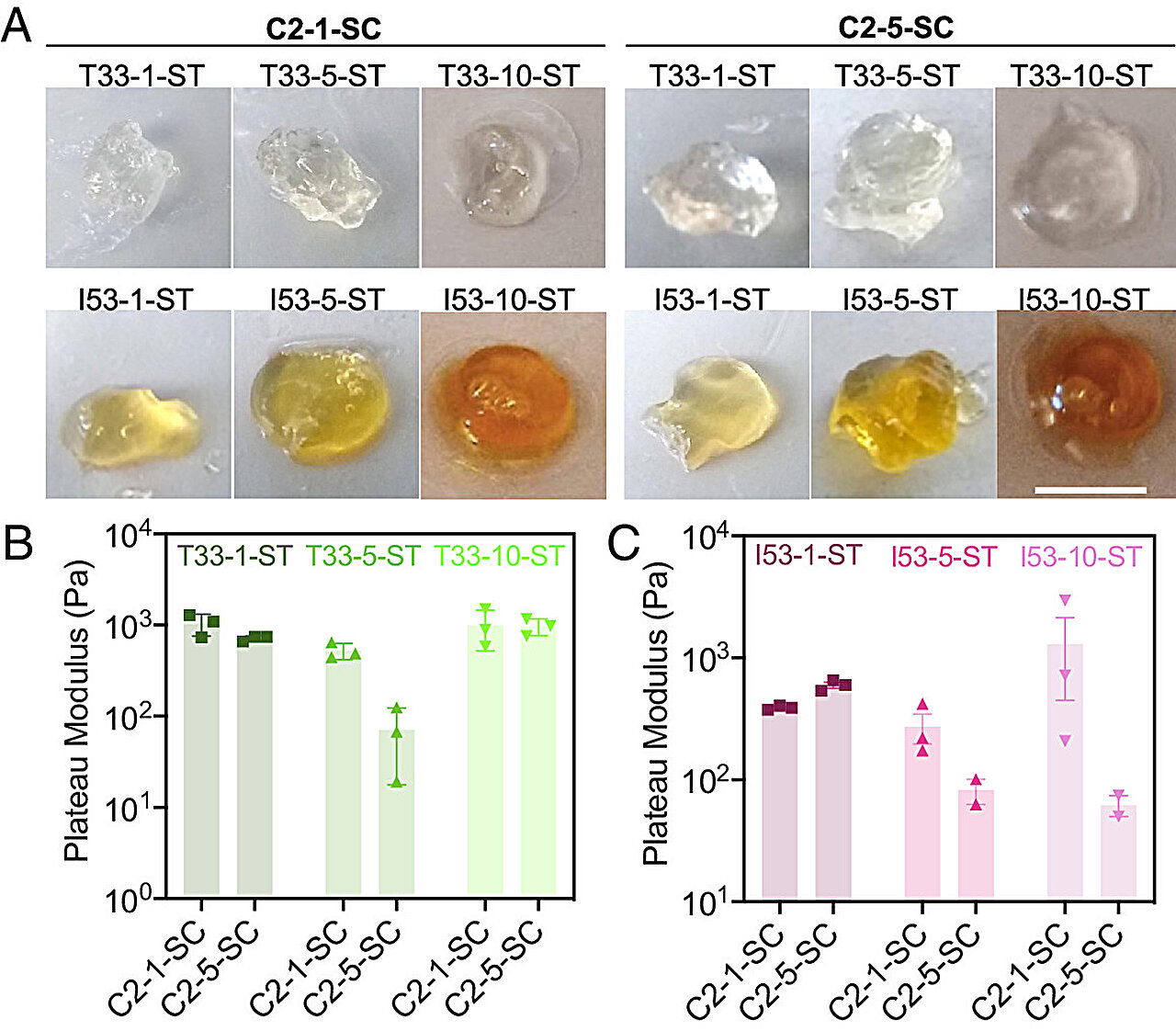
To determine if the hydrogels in cells had similar characteristics compared to their extracellular counterparts, the researchers examined whether building blocks within the hydrogels could move around. A stiffer hydrogel would be more likely to trap the proteins in one position compared to a more fluid gel. The mechanical properties of each type of hydrogel remained even when inside a cell.
The team plans to further explore this system, including being able to better control how hydrogels form and localize within cells.
The most crucial part of this project, the researchers said, was the collaboration between protein designers and chemical and biological engineers.
“Our cross-disciplinary collaboration with Cole’s group has been very exciting, and has opened up routes to new classes of biomaterials with a wide range of applications,” said co-senior author David Baker, the director of the Institute for Protein Design.
More information:
Rubul Mout et al, De novo design of modular protein hydrogels with programmable intra- and extracellular viscoelasticity, Proceedings of the National Academy of Sciences (2024). DOI: 10.1073/pnas.2309457121
Citation:
Computer-designed proteins allow for tunable hydrogels that can form both inside and outside of cells (2024, January 31)
retrieved 31 January 2024
from https://phys.org/news/2024-01-proteins-tunable-hydrogels-cells.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.